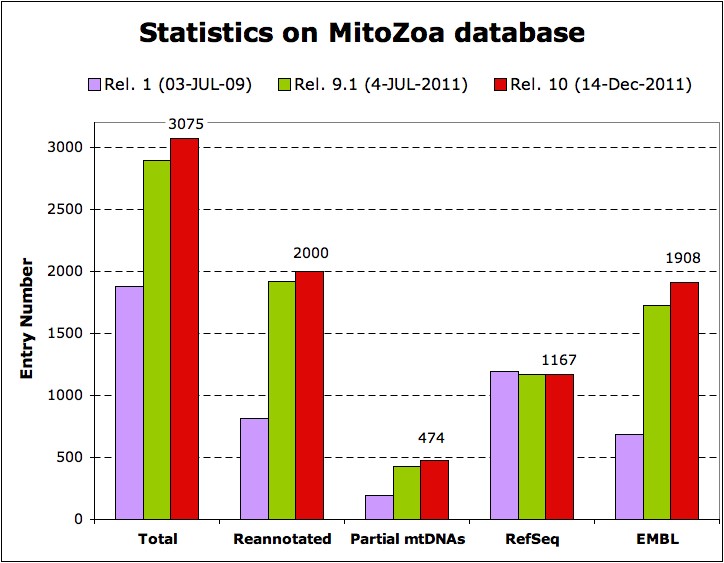
Statistics
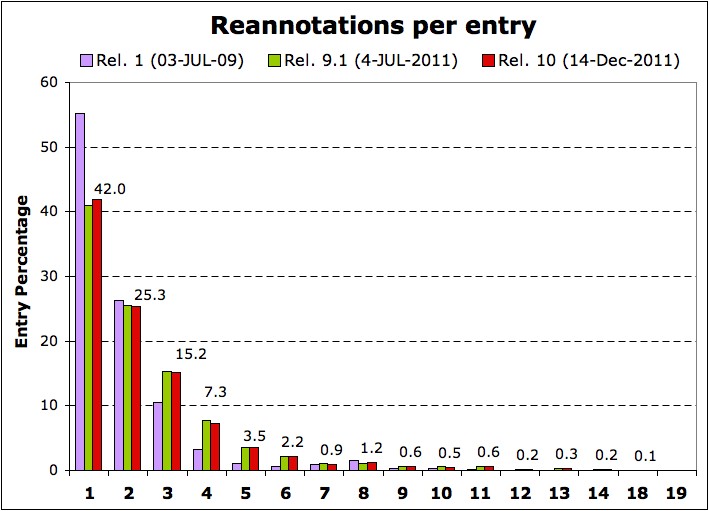
| Reannotation typology | Rel. 1 (03-JUL-09) | Rel. 8 (16-JUN-11) | Rel. 9.1 (04-JUL-11) | Rel. 10 (14-DEC-11) |
|---|---|---|---|---|
| Error in molecule topology | 173 | 374 | 402 | 438 |
| Modification of the Organism Species field | 6 | 8 | 8 | 20 |
| Modification of the DEscription field | 120 | 221 | 222 | 230 |
| Elimination of erroneous FTKey | 8 | 11 | 11 | 13 |
| Unannotated gene | 67 | 86 | 86 | 90 |
| Error in rRNA gene name | 7 | 4 | 4 | 4 |
| Error in tRNA gene name | 116 | 143 | 143 | 147 |
| Error in CDS gene name | 0 | 2 | 2 | 2 |
| Error in strand of rRNA | 12 | 34 | 34 | 37 |
| Error in strand of tRNA | 383 | 546 | 547 | 544 |
| Modification of tRNA anticodon specificity a | 994 | 2231 | 2280 | 2540 |
| Error in tRNA gene boundaries | 139 b | 2235 c | 2266 c | 2302 c |
| Error in rRNA gene boundaries | 10 | 13 | 15 | 19 |
| Error in CDS gene boundaries | 8 | 96 | 205 | 207 |
| tRNAs not validated d | 402 | 286 | 286 | 294 |
a: for tRNA genes decoding the same amino acid, i.e. trnG, trnL, trnM and trnS
b: only error in tRNA limits affecting gene order
c: any kind of erroneous tRNA limits, including those not affecting gene order
d: this number includes tRNAs not validated by the reannotation pipeline and short tRNAs (< 45 bp)
not checked by the pipeline due to their inability to form a complete secondary structure.
Taxonomical Statistics
Entry number of the main Metazoa taxa (Rel. 10, 14-DEC-11).| Taxon | Total | Partial | Linear | Congeneric | Mini-circle |
|---|---|---|---|---|---|
| Acanthocephala | 1 | 0 | 0 | 0 | 0 |
| Annelida | 18 | 9 | 0 | 0 | 0 |
| Arthropoda | 527 | 112 | 1 | 156 | 2 |
| Brachiopoda | 5 | 1 | 0 | 0 | 0 |
| Bryozoa | 3 | 0 | 0 | 0 | 0 |
| Cephalochordata | 9 | 0 | 0 | 8 | 0 |
| Chaetognatha | 5 | 0 | 0 | 0 | 0 |
| Cnidaria | 70 | 22 | 25 | 21 | 0 |
| Ctenophora | 1 | 0 | 0 | 0 | 0 |
| Echinodermata | 29 | 3 | 0 | 9 | 0 |
| Echiura | 2 | 0 | 0 | 2 | 0 |
| Entoprocta | 2 | 0 | 0 | 0 | 0 |
| Hemichordata | 3 | 0 | 0 | 2 | 0 |
| Hyperotreti | 2 | 0 | 0 | 0 | 0 |
| Mollusca | 148 | 14 | 0 | 74 | 0 |
| Myzostomida | 2 | 2 | 0 | 0 | 0 |
| Nematoda | 75 | 9 | 0 | 44 | 2 |
| Nemertea | 5 | 1 | 0 | 3 | 0 |
| Onychophora | 6 | 2 | 0 | 2 | 0 |
| Platyhelminthes | 45 | 5 | 0 | 34 | 0 |
| Porifera | 31 | 2 | 0 | 4 | 0 |
| Priapulida | 2 | 1 | 0 | 0 | 0 |
| Rotifera | 3 | 0 | 0 | 0 | 0 |
| Sipuncula | 3 | 1 | 0 | 0 | 0 |
| Tardigrada | 2 | 0 | 0 | 0 | 0 |
| Tunicata | 13 | 0 | 0 | 5 | 0 |
| Vertebrata | 2062 | 290 | 0 | 990 | 0 |
| Xenoturbellida | 1 | 0 | 0 | 0 | 0 |
| Total | 3075 | 474 | 26 | 1354 | 4 |
BLAST Database Statistics
Databases referred to Rel. 10 (14-DEC-2011).| Database | Sequences |
|---|---|
| mtDNA | 3075 |
| CDS_nt | 39341 |
| tRNA | 64528 |
| rRNA | 6058 |
| NCR >=25nt | 9377 |
| Protein | 39335 |
History
Warning on mt entries not included in MitoZoa: four entries have not been included in MitoZoa due to the lack of annotations of all genes and/or low sequence quality (AAPE02072785, FJ403244, HQ330989, HQ415764).
Novelties of Rel. 10: deletion of two duplicated entries (AP002931, NC_004449). Deletion of six entries retired by the Authors or by RefSeq (GU936203, GU936204, JF793665, NC_003170, NC_004383, NC_008943). Substitution of the entry NC_009082 with HM600781, due to uncertain gene annotation. Reannotation of entries belonging to Onychophora. Inclusion of three cnidarian entries shorter than 7 kb, because of their taxonomic importance.
Novelties of Rel. 9: optimization of reannotation of the protein-coding genes; annotation of "loss of highly conserved regions" in protein-coding genes, possibly due to frameshift or nucleotide substitutions.
Novelties of Rel. 8: implementation of a BLAST service against MitoZoa and against five distinct datasets of functionally homogeneous mitochondrial sequences; reannotation of protein-coding genes; identification and standardization of the annotation of protein-coding genes showing frameshift sites post-transcriptionally corrected by RNA editing or programmed translational frameshifting; creation of the new FTkey "prec_ORF" (precursor ORF) including a precursor ORF with frameshift(s) recovered by RNA editing or programmed translational frameshifting; updating of the "Organism Species" (OS) and "Organism Classification" (OC) fields of all existing MitoZoa entries; automated identification of gene order differences in congeneric species, and validation/correction through literature check; deletion of a duplicated entry (EF035448).
Correction in Rel. 7.1: modification of the anticodon specificity of five tRNA genes in entries EU345430, EU747728, EU682403, HM189212 and HM753535.
Novelties of Rel. 6.2: correction of a few bugs in the annotation of some NCR; annotation of all genes in three new entries (D83491, L07095, L07096), and completion of the annotation in two existing entries (AJ426041, AY250707).
Correction in Rel. 6: deletion of 24 duplicated entries (AB016274, AB079597, AB238966, AB242163, AF106038, AJ001562, AM181033, AM181034, AM181036, AP002929, AP004101, AP004355, AP004414, AP004417, AY158677, AY525783, DQ068951, DQ080041, DQ333813, DQ355300, DQ355301, EF071948, FJ590427, FJ752428).
Novelties of Rel. 5: modification of the identification criteria of the "longest non-coding region".
Corrections in Rel. 4.8: modification of annotations and comment lines in 10 entries, mostly belonging to congeneric species (AB360979, AB361005, DQ52643, DQ526431, DQ665851, EU266073, FJ529186, GQ265897, GQ888714, NC_009687), and deletion of the duplicated entry NC_006899.
Novelties of Rel. 4.7: annotation and name standardization of pseudogenes; inclusion of mtDNA entries > 7kb corresponding to subgenomic circles; update of the "Genetic Code" list; inclusion of additional information on gender-specific mitotypes; inclusion of additional information on RNA editing; improvement of the tRNA reannotation pipeline (with correction of small differences in tRNA limits); correction of bugs in rRNA reannotation; correction of bugs related to additional information on rRNAs.



